FTDNA now paints each chromosome according to ethnicity. That’s something 23andme has done for many years. How do they compare?
 The FTDNA site results are found under Results & Tools > Autosomal DNA > Chromosome Painting. You choose a colourful painted chromosome image, either Super Population or Continent. Also available are tabular detailed results.
The FTDNA site results are found under Results & Tools > Autosomal DNA > Chromosome Painting. You choose a colourful painted chromosome image, either Super Population or Continent. Also available are tabular detailed results.
In my case, the Continent results are all European, except for regions on chromosomes 1, 9, 13, 14, 15, 21 and 22 which are not assigned.
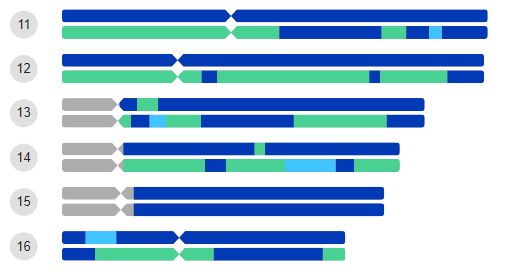
For Super Population my results, shown in part in the image, have segments for Western Europe, European Jewish (EJ) and Eastern Europe (EE). As I have all UK ancestry, except a Dutch Jewish line from my maternal grandfather, I was expecting to see one of each chromosome pair coloured for EJ ancestry, with maybe EE on the same chromosome.
My results had 11 instances where only one of the pair had EJ painted, sometimes with also some EE. 11 chromosomes were painted with EJ and/or EE on both sides. With EJ/EE on both chromosomes, one of each pair had the lion’s share. It appears the segments are not always properly assigned to the appropriate member of the chromosome pair.
I had 46 EJ segments in total.
In my results at 23andme only one of each pair was ever coloured for Ashkenazi Jewish (AJ_. Seven pairs showed no AJ. I had 32 AJ segments in total some of which bridged more than one EJ segment.
To bolster faith in the results both company’s analyses identified many of the same chromosome segments as Jewish.


They have an awful lot of work to do to make this tool useful for someone of just Western Europe descent. On their painter I’m about as mono-coloured as one could probably get.